Data Access Enhances Healthcare Research
August 23, 2021 | Susan Tussy
The National Institutes of Health (NIH) provide leadership and direction for research programs aimed at enhancing health, lengthening life, and reducing illness and disability. Data is an important ingredient for—and a product of—these projects. Historically, each program has managed the data it produces independently. Among the NIH institutes, a program called the Common Fund sponsors a portfolio of ambitious initiatives, each of which generates biomedical data of high value to the research community. Each Common Fund project focuses on a specific topic. Examples include cataloging the microbes that live in our bodies, the interaction between nerves and organs, pediatric cancer, and how chromosomes are organized in cell nuclei. The data-gathering component of each initiative is a team called a Data Coordinating Center (DCC), responsible for gathering and managing the data from participating projects and encouraging its use in new projects. Initiatives last up to ten years and they start and end on a rolling basis, with up to two dozen initiatives active at a time.
A notable feature of Common Fund initiatives is that they span a wide range of research disciplines, each with its own methods and conventions. For example, the data used to generate an atlas of the human body at the cellular level is significantly different from the data used to understand how chromosomes are organized in cell nuclei, which in turn is very different from the data used to understand how a child’s genome might be related to her pediatric cancer. Even something as fundamental as characterizing these datasets in a way that’s meaningful to researchers from a variety of disciplines is a challenge.
Common Fund Data Ecosytem
The Common Fund Data Ecosystem (CFDE) is one of several projects working to coordinate and enhance NIH’s management of its research data. CFDE is tackling that challenge, and others, with the goals of providing a unified inventory of the Common Fund’s data assets, ensuring those assets are maintained over time, and providing consistent mechanisms for researchers to gain access to datasets that can further their research.
While it isn’t hard to remember what kinds of data were collected, it turns out to be quite a challenge to remember enough about the data—how it was collected, where it came from, how each measurement was made, what permissions were given for its future use—for it to be useful in other research projects. There’s a lot of value in this data, but only if we remember enough about it for it to be reused. Otherwise, it’s just a very expensive pile of numbers.
FAIRer Data and Other Goals
One of CFDE’s specific goals is to encourage Common Fund DCCs to make their data FAIRer. FAIR[1] is an acronym that stands for “Findable, Accessible, Interoperable, and Reusable,” four qualities that improve the re-usability of research data. FAIRness is particularly important for Common Fund data because of the diversity of research methods used to produce the data and because of the wide range of biomedical disciplines in which the data might be used.
CFDE is working with the Common Fund DCCs to develop a common metadata model for Common Fund data: the CrossCut Metadata Model (C2M2) C2M2 provides DCCs with a way to describe their data holdings (the specific files and the relationships between files, research projects, species, anatomical tissues, and experiment types) that applies across all of the research disciplines and methods used.
Another of CFDE’s guiding principles is to leverage existing research and commercial services, minimizing CFDE’s development and maintenance costs. Though not limited to CFDE, NIH’s Science and Technology Research Infrastructure for Discovery, Experimentation and Sustainability (STRIDES) program is leading NIH-sponsored projects to use cloud storage platforms—such as Amazon’s S3, Google Cloud, and Microsoft Azure cloud storage—rather than developing their own proprietary storage systems. This simplifies data access for researchers and makes it easier for NIH to assume custodianship of the data as each DCC’s operational period ends.
The CFDE Web Portal
Summary View - Web Portal
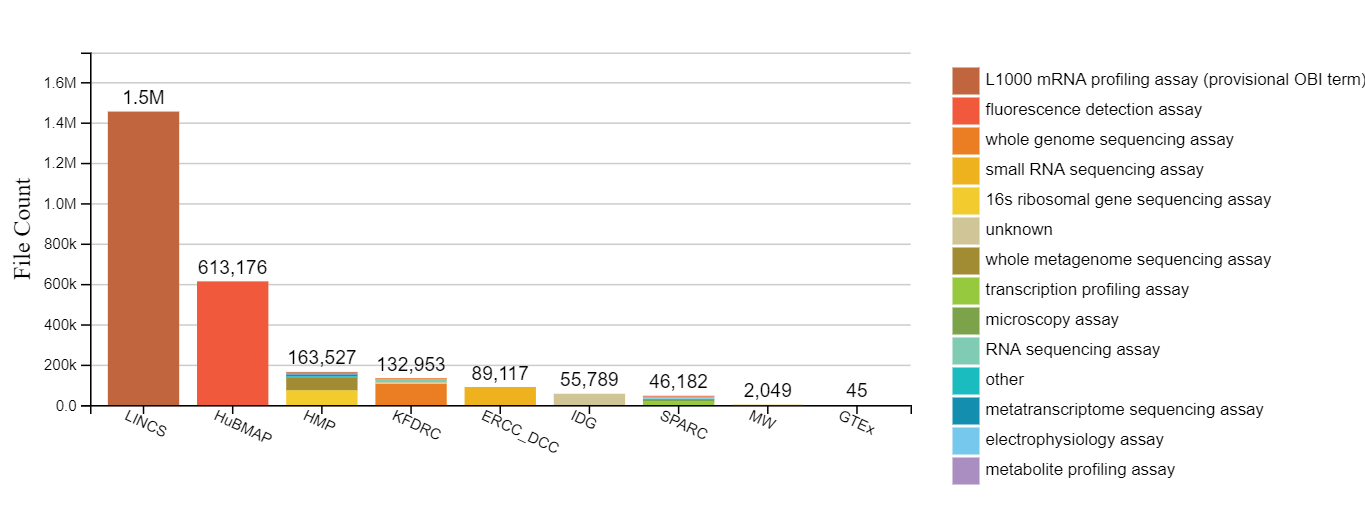
CFDE now provides a web portal that allows researchers to search and browse Common Fund data using the C2M2 descriptions provided by the Common Fund DCCs. The CFDE portal simplifies and automates many of the portal’s data management processes through the integration of the Flows service offered by Globus, an initiative out of the University of Chicago for the research community. Submissions are initiated by DCC personnel who have locally installed the cfde-submit CLI tool developed by the University of Chicago. Each submission is a complete inventory of a DCC's data holdings, including a complete file listing and research-relevant metadata. CFDE's submission Flow uses a Globus Connect Server-hosted collection and either Globus Connect Personal (GCP) or HTTPS upload to get the data off the submitter's local system and into CFDE’s AWS-hosted system for subsequent processing and loading for review & approval. Access control in the CFDE Portal is managed using Globus Groups. Currently there are eleven participating DCCs. Collectively, they manage over 800 projects, containing information on more than 650,000 bio samples collected from over 18,500 subjects from humans and organisms.
“Access to high-quality, curated data is a critical requirement for research in the life sciences,” states Ian Foster, Arthur Holly Compton Distinguished Service Professor, University of Chicago and Contributor to the Common Fund Data Ecosystem. “The CFDE portal makes datasets from multiple programs more FAIR (findable, accessible, interoperable, and reusable), while implementing any necessary security and access controls. The CFDE initiative and it’s data portal deliver important elements for accelerating discovery as we tackle important healthcare issues to improve people’s lives.”
Further integration with the Globus platform is underway which will enhance the portal’s capabilities. Engineers are beginning to connect the CFDE portal to NIH’s Researcher Auth Service (RAS), so researchers will be able to have insight into their current permissions in regard to controlled-access datasets.
Researchers will also be able to watch for relevant data and discover datasets related to specific genes, biomolecules and diseases.They will be able to follow their favorite data types, and share data amongst participating DCCs, third-party viewers, and analysis platforms.
As we move to more interdisciplinary research the need for FAIRer datasets will become more important than ever before.
References
[1] Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, IJsbrand Jan; Appleton, Gabrielle; et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship". Scientific Data. 3: 160018. doi:10.1038/sdata.2016.18